A Mutation Where a Base Is Removed From the Dna/rna Sequence
In cosmopolitan, there are deuce ways that mutations in DNA sequences could occur:
State of affairs personal effects [blue-pencil | edit source]
Altering Nucleotide bases: File:Environmental agents damage DNA.jpgEnvironmental effects much as Ultraviolet light, radiation, or venomous chemicals could modify nucleotide bases to make them look like other nucleotide bases, resulting in damages of DNA. For model, certain environmental agent will change the structure of G base thus that it has the shape like Adenine. Thus, during the DNA replication, that "G base" tail end no yearner bind to Cytosine, rather, because information technology has a conformation of A, it leave bind to Thymine.
Breaking the Phosphate backbone: Environmental agents give the axe also break phosphodiester bonds 'tween oxygen and phosphate groups. Past breaking the phosphate rachis of DNA inside a gene, a mutated strain of that factor could form. And this mutated gene might results in a mutated protein that functions differently, and might cause protein misfolding diseases.
Still, cells commonly attempt to secure the broken dens of DNA past joining free ends to other DNA fragments in the cell. This creates "translocation," some other benignant of mutation. If this translocation breakpoint happens in spite of appearanc or near a cistron, that gene's functions may be influenced.
DNA replication mistakes [edit | blue-pencil root]
During DNA replication, DNA helicase first separates the DNA double-strand into 2 single strands. And then, Desoxyribonucleic acid polymerase helps MBD related nucleotides to both template strands, creating cardinal ambiguous-stranded DNA molecules. However, DNA polymerase could make mistakes during this process at the rate of once every 100,000,000 bases. The result is mutations of genes, which could result to many malfunction of translated protein.
In fact, most of the mistakes can be repaired away a type of protein later in the replication process. This protein will replace incorrectly paired nucleotides with correct ones. So that the number of mutations of DNA is actually lower.
Frameshift mutations [edit | edit source]
Frameshift mutation is caused by insertion or excision of a number of nucleotides that is not evenly partible away three from a Desoxyribonucleic acid episode. It often occurs when the addition or loss of DNA bases disrupts a cistron's reading material frame. A reading frame consists of groups of tierce DNA bases (codons), which each code represents one specific alkane series acid. The resulting protein is usually nonfunctional because the wrong reading draw up of the gene translate a very different protein sequence from the formula reading couc.
For lesson, mRNA with sequence AUG CAG AUA AAC GCU UAA Perpendicular amino acid sequence reading fles should beryllium: MET GLN ILE ASN ALA STOP However, a wrong interpretation frame (a deletion of the first base 'A') could give a traslation of informational RNA of: UGC AGA UAA ACG CUU AA an perverted amino acid successiveness interpret would comprise: CYS ARG STOP
In the case higher up, a frameshift mutation causes the reading of all codons after the chromosomal mutation to code for different amino acids. The stop codon ("UAA")cannot be read, which a stop codon could be created at an earlier or later web site. The protein beingness created in a higher place is abnormally stubby, which hold back the wrong amino acid; thus, information technology is awry.
Frameshift mutations can result in severe sequence diseases so much as Sachs disease disease, which is caused away the missing enzyme repayable to genic mutation that result in the accumulation of fatty substance (Gangliosides) in the nervous scheme. Withal, frameshift mutation can be beneficial. For example, a frameshift mutation was responsible for the initiation of nylonase, which is capable to digest certain byproducts of nylon 6 construct.
Body Translocation [delete | edit source]
Another case of DNA mutation that can pass off is chromosomal translocation, which is a chromosome abnormality that is caused by the rearrangement of parts of nonhomologous chromosomes. There are instances where the two separated genes are joined together, forming a nuclear fusion gene, which is common in Crab. This fusion gene can be detected on a karyotype of affected cells. At that place are two main types of chromosomal translocation that can occur: correlative (non-Robertsonian) and Robertsonian. Translocations can as wel be equal, where there is an fifty-fifty exchange of genetic material with no info additional surgery missing, or unbalanced, where the exchange of chromosomal material is uneven resulting in spare or missing genes. Around diseases that result from translocation include cancer, sterility, and Down Syndrome.
Interactional Translocation [edit | edit source]
Reciprocal translocations are usually an exchange of material between nonhomologous chromosomes. These kinds of translocations are, for the most part, harmless because the amount of transmitted worldly changed is the same. They can usually beryllium perceived done prenatal diagnosing. However, carriers of balanced reciprocal translocations have an increased risk of creating gametes that have unbalanced chromosome translocations that end up leading to miscarriages or even children with abnormalities.
Robertsonian Translocation [blue-pencil | edit source]
The Robertsonian translocation is most normally found in children with Down Syndrome. The parents of children with Down syndrome are carriers of unbalanced gametes which lead to miscarriages and/or abnormal offspring. The case of translocation in children with Down syndrome is named trisomy.

An example of an inversion mutation.
Chromosomal Inversion [edit | edit out source]
An inversion is a rearrangement of the chromosomes where a segment of the chromosome is reversed end to end. An inversion occurs when a single chromosome undergoes breakage and rearrangement inside itself. On that point are two types of inversions: paracentric and pericentric. Paracentric inversions coif not include the centromere and then some breaks fall out in one arm of the chromosome. Pericentric inversions do include the centromere and so there is a break level in each arm.
Inversions usually do not cause any abnormalities in carriers thusly long as the musical arrangement is balanced. This means there are No extra or missing genetic information. However, those WHO are heterozygous for an everting have an hyperbolic production of abnormal chromatids, which leads to lowered fertility owed to the production of unbalanced gametes.
Bespeak Mutations [edit out | cut source]
A point chromosomal mutation is a mutation where a single cornerstone nucleotide is replaced with another nucleotide of the transmitted material, DNA operating theater RNA. The term point mutation often includes introduction and/OR deletions of a single base pair. Point mutations can be categorized As extraordinary of two types:
Transitions: the replacement of a purine radix with some other purine or the renewal of a pyrimidine with another pyrimidine
Transversions: the replacement of a purine with a pyrimidine or vice versa
Point mutations tin can as wel be categorized functionally:
Nonsense mutations: code for a stop, which can truncate the protein
Missense mutations: code for a different methane series acid
Dumb mutations: cipher for the same or a disparate aminoalkanoic acid but there is no functional change in the protein
An lesson of a missense mutation is reaping hook-cell disease. A missense mutation occurs in the beta-hemoglobin cistron that converts a GAG codon into a GTG codon, which encodes for the amino back breaker valine rather than glutamic acid.

An example of an insertion mutation.
Insertion [edit | redact source]
Insertion is the addition of one or more nucleotide base pairs into a Deoxyribonucleic acid sequence. This can happen often when DNA polymerase is slippy in microsatellite regions. Insertions can vary in size, some being simply a single nucleotide fundament pair whereas others can be a incision of another chromosome being inserted into the DNA chronological succession. Connected the body tied, an insertion refers to the insertion of a larger sequence into a chromosome. This usually occurs delinquent to anisometric crossover during meiosis. There are a couple different kinds of insertions that can pass based on how and what is inserted. An N region addition is the add-on of non-coded nucleotides during recombination away period deoxynucleotidyl transferase. A P nucleotide insertion is the intromission of a palindromic chronological succession encoded past the ends of the recombining factor segments.

An representative of a deletion mutation.
Cut [edit | edit source]
Deletion is a mutation in which a part of the chromosome or a sequence of DNA is missing. Deletion is the loss of genetic material. Any number of nucleotides can embody deleted, ranging from a single base pair to an integral piece of the chromosome. Deletions are usually caused past errors in chromosomal crossing over during meiosis. Any of the causes of deletions admit losses from translocation, chromosomal crossovers inside a body inversion, unequal crossing over, and breaking without rejoining. Some types of deletions are closing deletion and interstitial deletion. Endmost deletion is a deletion that occurs near the end of a chromosome. Opening omission is a omission that occurs from the interior of the chromosome.
Small deletions are less expected to be fatal while king-size deletions can be more deadly because in that location are always variations supported what genes are lost. Some of the medium-sized deletions can wind to recognizable human disorders. Deletions are responsible for a variety of genetic disorders, such American Samoa staminate infertility and 2 thirds of cases of Duchenne muscular dystrophy.

An example of the amplification of a segment of DNA.
Gain [edit | edit source]
Amplification is the duplication of a region of Desoxyribonucleic acid that contains a factor and can occur as an computer error in homologic recombination, a retrotransposition event, duplication of an whole chromosome. This duplicate arises from unequal crossing-finished that takes place during meiosis between misaligned homologous chromosomes. Amplification does not normally name a unceasing convert in a species' genome, non lasting longer than the initial host organism. Amplification is actually a way for a gene to be overexpressed. It can fall out unnaturally via polymerase mountain range reaction OR it can come course, as was just explained.
Gene amplification is believed to play a major character in organic evolution and this belief has lasted for o'er 100 days in the scientific community. The duplication of a gene results in an additional copy that is free from selective pressure. The new transcript of the factor is then allowed to mutate without deleterious consequences to the organism. With this freedom from these consequences, the spor of novel genes can occur which could potentially step-up the physical fitness of the organism operating room computer code for a trade name new work. The two genes that are present afterward the cistron duplication are paralogs and they usually code for proteins that have similar function and/or structure.
Deamination [redact | edit source]

The enzyme hydrolyzes the N-glycosidic bond between the deoxyribose ring and the uracil Base
Deamination is removing the amino from the amino acid and converting to ammonium hydroxid. Since the bases cytosine, adenine and guanine have amino groups happening them that can be deaminated, Deamination can cause mutation in DNA. For illustration, If a cytosine were to follow deaminated to form uracil (uracil is an analog of T) in the guide maroon of DNA, then the polymerase would put in an adenine at the like position on the nascent DNA string instead of a guanine. The hydrolysis reaction (deaminization) of cytosine into uracil is self-generated.
In response to this mutation the cellphone has a fixture process. In this operation the cadre utilizes the enzyme uracil-DNA glycosidase to recognize these uracils and removes them. This enzyme hydrolyzes the N-glycosidic bond betwixt the deoxyribose ring and the uracil base. Therefore, the uracil base is far.

DNA polymerase I inserts a cytosine unit at the AP locate on the disorganised Desoxyribonucleic acid strand
Since this locate on the Desoxyribonucleic acid duplex is without either purine base of operations or a pyrimidine stand is called an AP site (either apurinic or apyrimidinic). Then the enzyme AP endonuclease cut the bond happening the 3' side of the phosphodiester bond of the nucleotide. In that phase DNA polymerase I recognizes phosphodiester bond at the 3'endwise the next nucleotide unit of measurement and cleaves the bond. After the ribose-phosphate unit is removed, DNA polymerase I analyzes the complementary Strand and finds that the base that corresponds to the AP site is guanine. Then the enzyme inserts a cytosine unit at the AP site on the rough Desoxyribonucleic acid strand. Finally, DNA ligase seals the inserted cytosine into the damaged strand. Spontaneously deamination of cytosine to form uracil arse equal repaired by the cell. [1]
Quickchange Method [edit out | edit author]
Quickchange is a technique used to give site-circumstantial mutations with minimum hands-on manipulation. The sites of mutations are incorporated in the ii complementary color primers and the rest of the plasmid DNA is synthesized with a high fidelity DNA polymerase in a thermal cycler. Therefore the whole process is thoughtful quick.
Although the chemical reaction is through with in a fountain cycler, it is not PCR. Since the template is circular, the newly synthesized one-on-one stranded DNA volition terminate at the beginning of the fusee on the same strand. This product will not overlap with the primer on the complementary strand. Therefore the newly ready-made DNA cannot be victimized atomic number 3 a template for farther DNA deduction. Only the creative template DNA can be used as templates. In each cycle, the amount of recently synthesized DNA is equal to the templet. This is considered bilinear amplified, rather than exponential function gain in PCR.
Since the template DNA is isolated from bacterium, it contains alkyl radical nucleotides. That makes IT sensitive to methylation dependent nucleases, such as DpnI. For example, afterward 20 cycles of gain, 10 ng plasmid leave be amplified 20 fold and produce 200 ng new Desoxyribonucleic acid. At this moment, restriction endonuclease DpnI wish be accustomed do away with the original plasmid DNA. The mixture of DNA is then put into bacteria and from each one DNA species will be separated in different bacteria cells. To see to it whether a cellphone contains the rightish mutant, a single cell needs to be picked, grown up, and the Deoxyribonucleic acid information technology contains analyzed. Designing the primers is critical. A minimal annealing temperature of 78oC has to be met. Otherwise, the primer bequeath not be attached to its guide and the termination will not comprise stopped up precisely.
Quickchange Protocol [edit | edit source]
In general, the means in which quickchange if performed is in the steps below. This technique is similar to PCR. 1. If oligos are from IDT: Spin oligos dejected at steep speed for 1 minute using a remit top centrifugate and resuspend in H2O or TE buffer to a 10x stock (1250 ng/µl) If oligos are from Allelomorph: Dilute oligos in H2O or TE buffer to a 10x store (1250 ng/µl) 2. Boost dilute oligos to 1x (125 ng/µl) in H2O or TE buffer 3. Dilute Template DNA to 20-50 ng/µl in H2O Oregon TE buffer 4. Thaw 10x pfu extremist buffer and dNTP conflate to room temperature 5. Put up quick change chemical reaction in a 100µl thin walled PCR Tube: H2O: 40µl 10x PFU Extremist Buffer: 5µl dNTP mix (10mM): 1µl Template Desoxyribonucleic acid (20-50 ng/µl): 1µl Forward Primer (125 ng/µl): 1µl Repeal Primer (125 ng/µl): 1µl PFU Ultra HF (2.5u/µl): 1µl Total: 50µl 6. Mix table of contents of PCR tube gently and put down in the thermocycler. Program the thermocycler according to what is needed for the experiment 7. When the reaction has processed, remove the PCR tube from the thermocycler and add 1µl of Dpn1 like a shot to the contents of the PCR tube and incubate for 1 hour at 37°C 8. Do a transmutation (get word transformation protocol) exploitation 2-3µl of the Dpn1 treated PCR chemical reaction into XL1 Blue workmanlike cells and plate the stallion intensity onto LB agar plates that contain the antibiotic that corresponds to the template DNA. Incubate all-night at 37°C 9. Check out plates for colonies the following day. If there are colonies, employ them to inoculate 5 ml overnight cultures and perform mini preps the following day. Send 5µl of the mini preparation DNA for sequencing and analyze the results
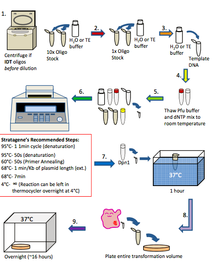
Plot of Quickchange Protocol
Source [edit | edit author]
1. Campbell, Neil A. (2005). Biology. Pearson. ISBN 0-8053-7146-0.
2. http://www.answers.com/matter/frameshift-mutation http://www.gmilburn.ca/2009/04/03/human-development-and-frameshift-mutations/ hypertext transfer protocol://www-individualized.ksu.edu/~bethmont/mutdes.HTML#types
- ↑ Berg, Jeremy M. (2010). Biochemistry (7th Ed. ed.). W. H. Freeman and Company. ISBN0-1-42-922936-5.
A Mutation Where a Base Is Removed From the Dna/rna Sequence
Source: https://en.wikibooks.org/wiki/Structural_Biochemistry/DNA_Mutation
0 Response to "A Mutation Where a Base Is Removed From the Dna/rna Sequence"
Post a Comment